Force Field
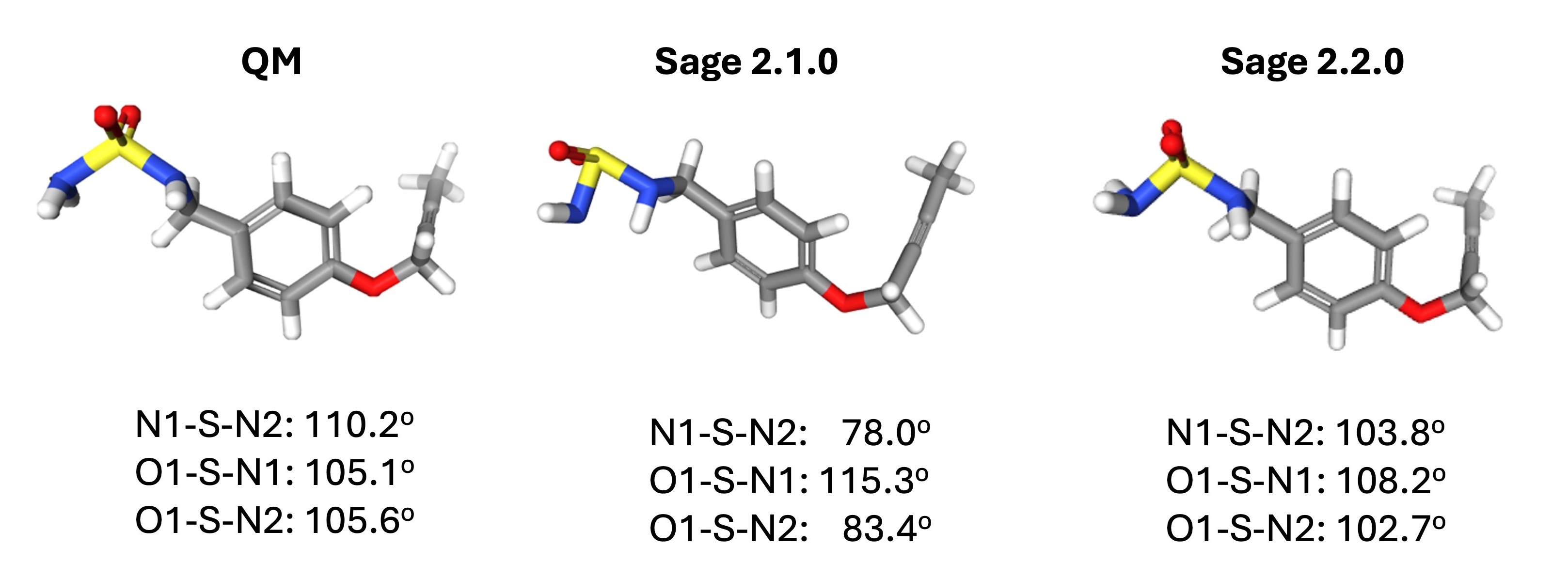
Improvements coming in the Sage 2.2.0 force fieldPosted on 26 Mar 2024
by Alexandra McIsaac
We are excited to share the release of our first release candidate of our newest force field, Sage 2.2.0!
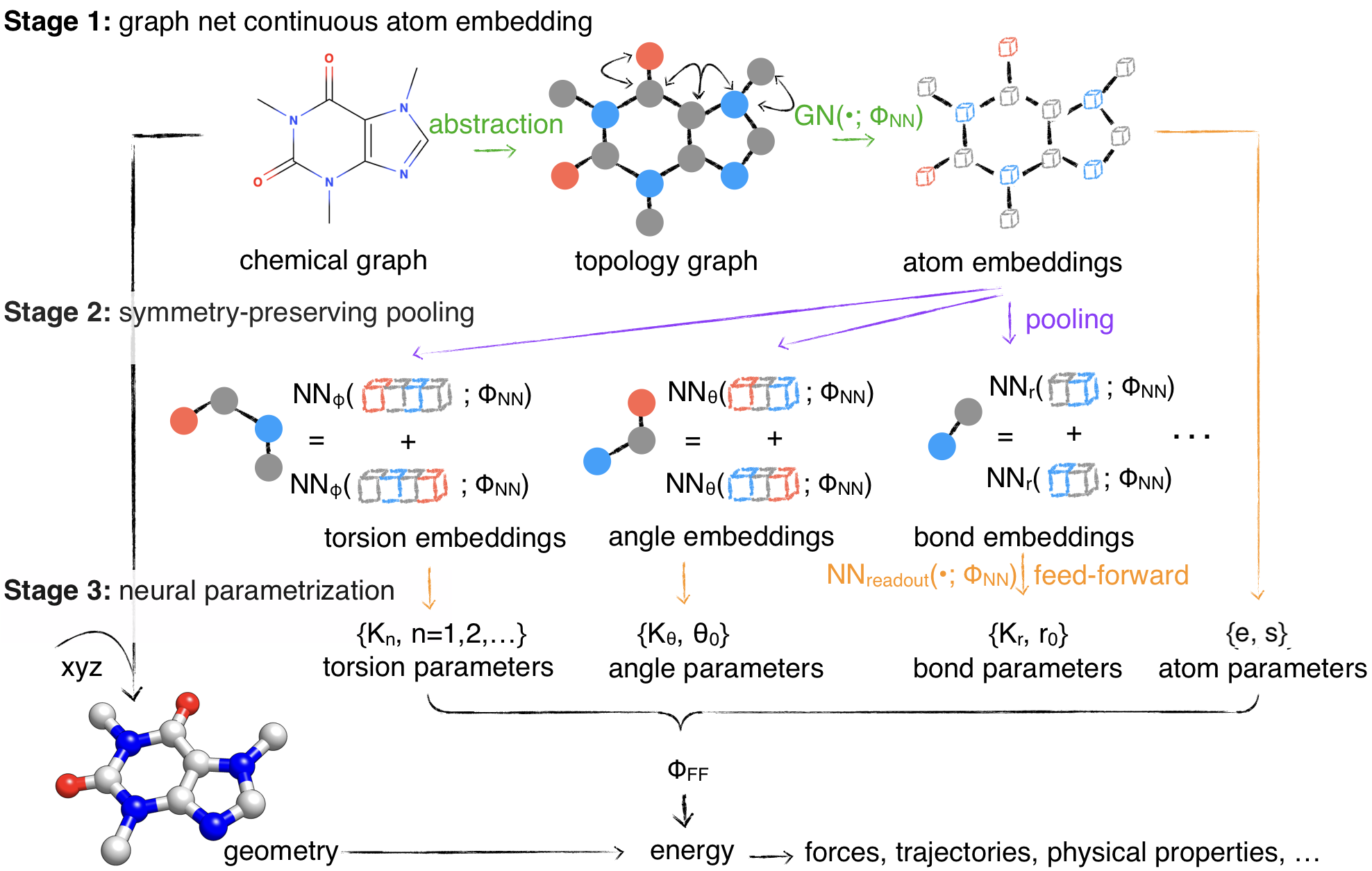
Self-consistently simulating small molecules, proteins and RNAs with the "espaloma-0.3" force fieldPosted on 12 Feb 2024
by Yuanqing Wang
I want to get out of building and improving force fieldsPosted on 31 Oct 2023
by David Mobley
It might seem odd for me to say this, because I’m one of the principal investigators helping to lead the Open Force Field Initiative. However, I think many of us involved in the project actually feel the same way.
Open SciencePosted on 19 Dec 2022
by Diego Nolasco
By working together, we can make this happen
Training force field parameters to Host-Guest bindingPosted on 31 Aug 2022
by Jeffry Setiadi
Tuning GBSA parameters to host-guest binding data using Open Force Field
Open Source, Enterprise-level Data PipelinePosted on 10 Jun 2022
by Pavan Behara
Molecular dataset generation workflow
Benchmark: Collaborative assessment of molecular geometries and energies from the OpenFFPosted on 15 Apr 2022
by Lorenzo D'Amore
Assessment of molecular geometries
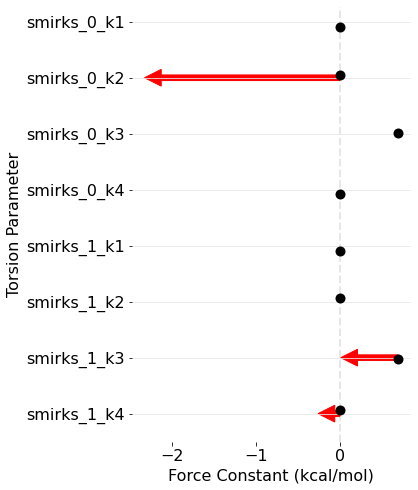
How to train your force field 2: Bespoke fitPosted on 20 Oct 2021
by Joshua Horton
Generating bespoke torsion parameters on the fly.
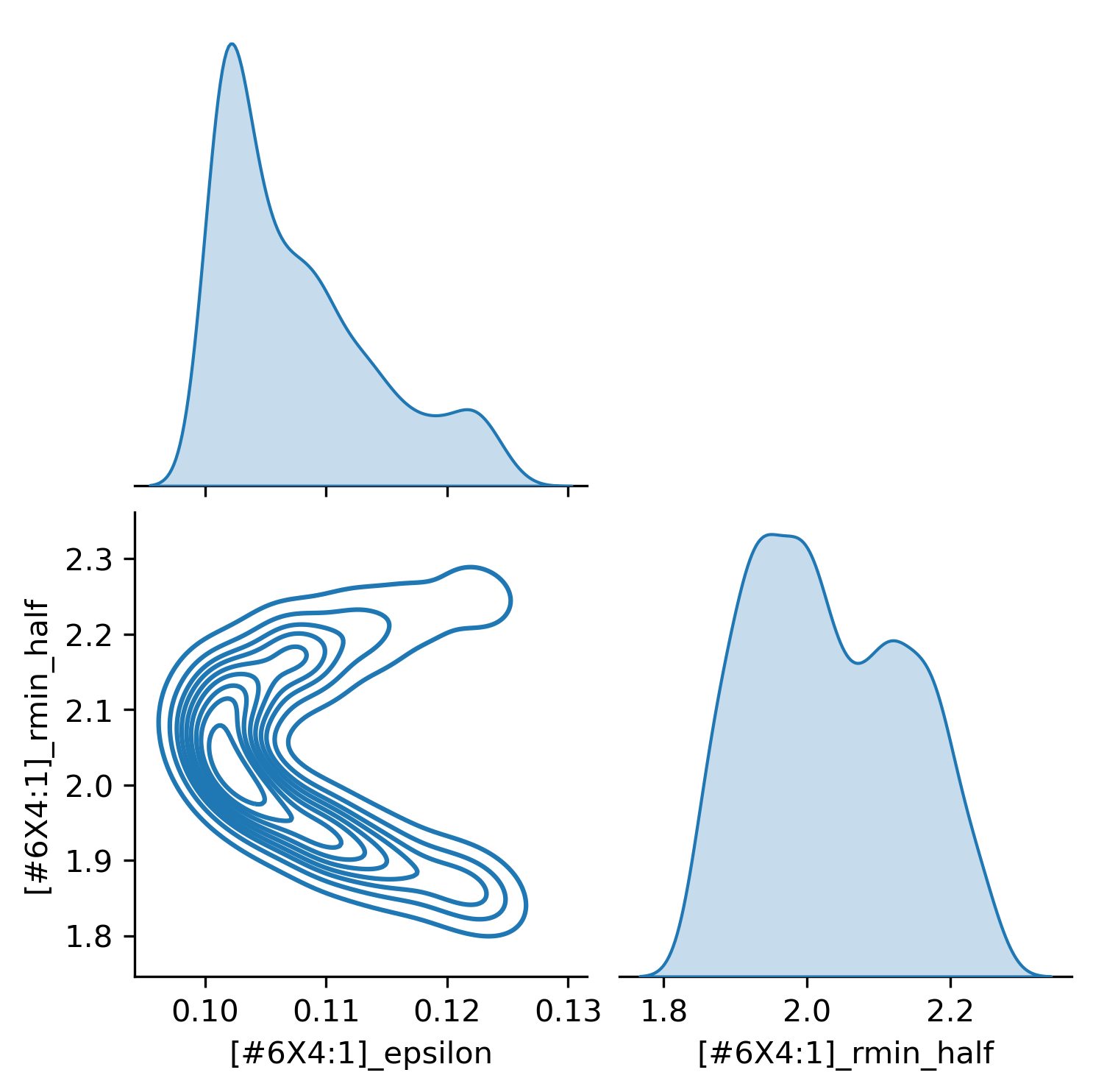
Surrogate-enabled Bayesian sampling of force field parametersPosted on 3 Sep 2021
by Owen Madin
Building and sampling surrogate models of physical properties in the OpenFF workflow
1 of 2
Next Page
