Release-1 update (Aug 15, 2019)
Posted on 20 Aug 2019 by Karmen Condic-Jurkic
Chemical coverage
C. Bayly suggested broadly dividing parameters into two categories: 1) chemistry being covered in the first optimization run; 2) everything else. The latter category should also include information about all molecules that were cleaned out from databases used in the first optimization sprint, which was focused on improvement of a selected (sub)set of parameters. This information (data) can be used in the later stages of the force field optimization when chemical coverage will be expanded. J. Wagner will create a new repository that will host conda installable new force field, which also seems like the best home for the chemical coverage information. In the meantime, this data will reside in QCA submissions repo.
D. Mobley raise the issue of introducing new parameters in the later phases of optimization, namely, after LJ parameters enter optimization cycle. Y. Qiu believe that he will have bandwidth to run additional valence optimizations, but everyone agrees that any parameters being used in LJ optimization should not be changed in these additional valence optimization steps due to high computational cost of LJ parameterization procedure.
LJ (vdW) fitting
S. Boothroyd gave an update on PropertyEstimator (PE) and ForceBalance (FB) interface and initial fitting tests. All tools and documentation around decisions made in building the initial datasets for Lennard-Jones parameter optimization can be found in a GitHub repository called nistdataselection. The initial set is on the small side, but it’s a good starting point. Initial tests for using PE and FB to optimize LJ parameters, requiring ~ 30 simulations, were successful and improved the objective function. The tests have been now expanded to a larger set containing ~60 simulations. These tests were run only with densities and heats of vaporization at the moment. S. Boothroyd argued against fitting to dielectric constants at this stage due to unfavourable ratio of computational cost vs parameter improvement (dielectrics require 10x more expensive computations to achieve only minor changes in LJ parameters). Additionally, the gradient of the dielectric constant with respect to possible parameters seem to be incredibly noisy and he worries that optimizing against this amount of noise could be potentially harmful. He advised to start using dielectric constants after we better understand the effect on the parameters and value of including this particular property in fitting. L.-P. Wang agreed that it’s harder to tell if the optimizer is working correctly with a lot of noise in gradients, D. Mobley also mentioned the role of electrostatics in predicting dielectric constants and everyone agreed to leave dielectrics out of optimization cycle for now due to tight timelines, and until changing electrostatics become a part of fitting.
At the moment, the dataset used (larger test set) includes at least 2 density and 2 Hvap data points for each SMIRKS pattern, and about 15 LJ terms are being optimized. This selection approach covers a lot of basic chemistry, including carbons and halogens (but not iodine), but ionic systems and some weird chemistry is not picked up. The initial parameters used are from 0.0.7 version of the force field (link). S. Boothroyd is to check how much LJ parameters change during optimization. All the simulations are being run with H-bond constraints.
Torsion/LJ fitting: . Boothroyd raised the question of (de)coupling LJ and torsion fitting and the interdependence of these parameters and how to approach co-optimization. C. Bayly explained that from the legacy point of view, the vdW terms were kept fixed and they would never allowed to change as a result of torsion information, but torsion information would changes as a result of vdW terms. He advised some caution in co-optimization, if some torsion terms would end up affecting vdW terms. In general, finding the best torsions that work with a given set of frozen vdW parameters should be a good strategy for now.
Valence/torsion fitting
Detailed documentation of all the existing optimization rounds of valence parameters can be found here: https://github.com/lpwgroup/forcebalance-qcarchive/releases. In this meeting, the latest versions (v.0.0.7 and v.0.0.8) were discussed. The previous release v.0.0.6 included vibrational frequency in fitting and three parameters resulted with a large change of force constants due to the large initial value, which were then modified in v.0.6.1. The version v.0.0.7 includes new torsion profile targets, as suggested by C. Bayly, and the results are encouraging. The new torsion profile targets includes torsional fitting with relaxed MM energies (see figure below). This release is complemented by detailed analysis by H. Jang of which parameters are improved and which are made worse during the optimization.
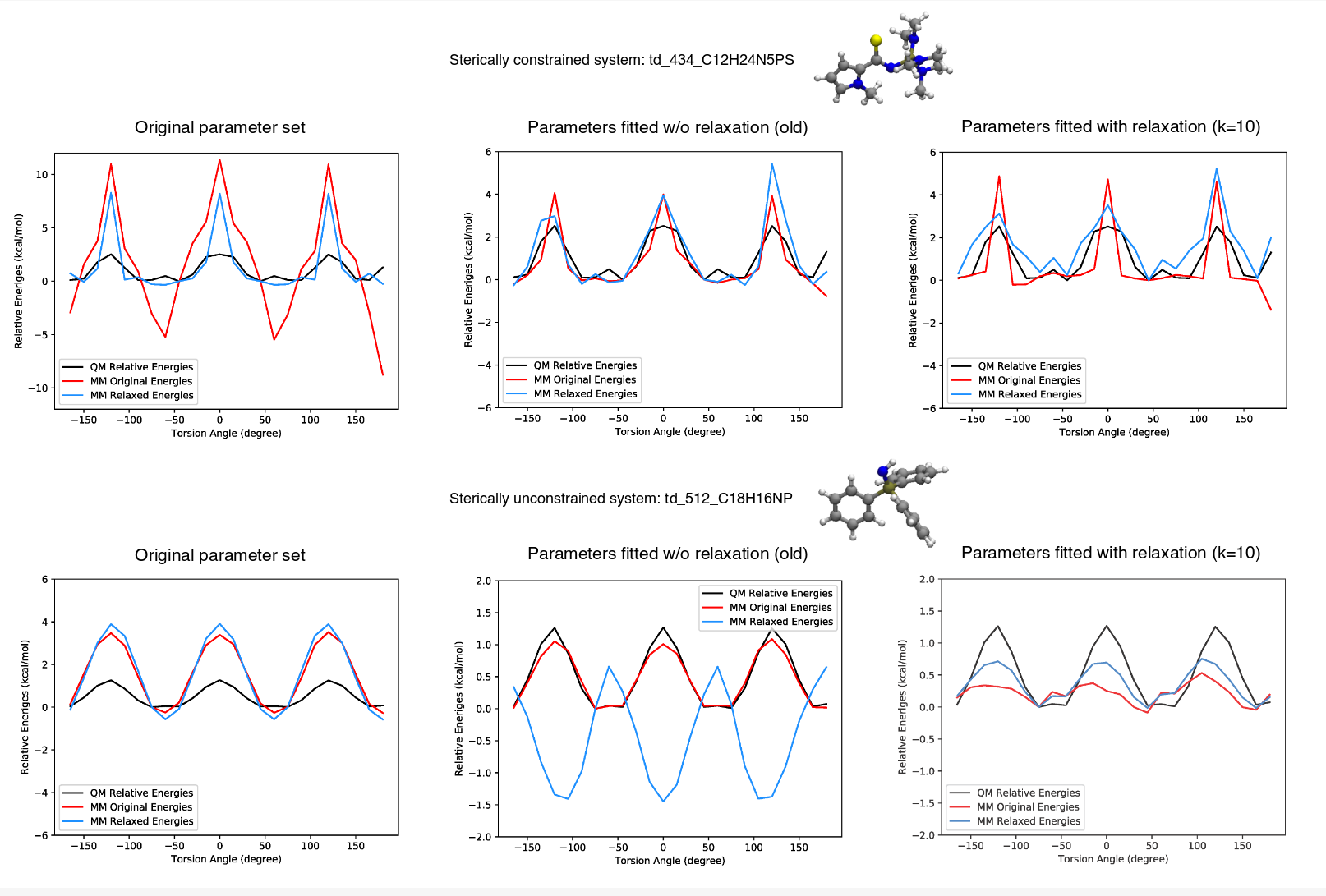
Version v.0.0.8 is pretty similar to the previous release, but it contains results for the valence fitting with new torsion term “t155b”, which allows using more QM data as the fitting target (~60-70 more molecules). Y. Qiu further noticed that OpenMM energy minimizer doesn’t necessarily result with the same geometries between different fitting rounds and he tried to address this issues in v.0.0.8.1. The latest v.0.0.8.2 (https://github.com/lpwgroup/forcebalance-qcarchive/releases/tag/v0.0.8.2) includes fitting with certain linear angles that were kept fixed at 180°.
Problematic cases: H. Jang investigated whether ELF method can be used to identify conformers with strong intramolecular interactions before running torsion scans and analysed the largest contributions of destabilizing Coulomb energy. It is not clear whether all the molecules that were flagged as potentially problematic for torsion scans using this approach should be excluded from torsion fitting. C. Bayly commented that the presence of strong intramolecular interactions could be more problematic in AM1-BCC fitting, but not necessarily for torsions. The main conclusion is that more research is required to come up with some reliable heuristics to use for flagging potentially problematic molecules before they get included for torsion scans/fitting.
Isothiocyanate (N=S=O) moiety has been recongised as problematic, where the angle has changed from 180° to 147° in some of the fitting rounds. The generic a34 parameter assigned may be too general and covered the unwanted chemistry, which will be fixed by adding some new terms in the next release (potentially a good case for Chemper).
Data handling
A new repository on Github will be created for S. Boothroyd results, similar to Y. Qiu’s ForceBalance-qcarchive repository for valence terms. This repository will also be the place where the final data used in the first optimization sprint will be stored. In any case, the information from both repos will have to be merged.
Validation set
C. Bayly suggested to create a set of evaluation molecules with certain geometries for which associated energies exist to validate departure from the canonical Smirnoff99frosst, and from release to release. This validation test should be a part of each release. This molecule set can also be used as a test in the future to see if any molecules are inadvertently changing with the force field update, and it should exercise each parameter at least once. D. Mobley suggested using the coverage set for this purpose, and L.-P. Wang suggested single point MM energies associated with geometries that correspond to the minimum as well as the ones away from it. The exact details for implementation and computational details of this kind of validation test need further discussion to ensure reproducibility by other users for the information provided, but there is a general agreement that this would be a valuable addition to every information. Y. Qiu will try to come up with an initial setup using coverage set.
