Torsion Datasets and Benchmarking Update
Posted on 16 May 2019 by Karmen Condic-Jurkic & Lee-Ping Wang
Dataset
A set of 486 small and chemically diverse molecules, provided by Roche, was chosen for the initial scanning of torsions to generate relevant QM datasets for subsequent force field parameterization. This step is also meant to serve as an infrastructure test for each component of the torsion pipeline and to perform a benchmarking study for the most appropriate level of theory to be used in the future calculations of torsion profiles.
Molecules in the “Roche set” have less than 30 heavy atoms, which removed the need for fragmentation prior to torsion calculations in this instance. From the entire torsional space spanned by these molecules, a minimal set of 820 1D torsions were identified based on the following criteria:
- The torsion should involve four unique heavy atoms;
- The center bond should not be in a ring;
- Only one torsion per center bond (choose the largest groups as terminal atoms).
This initial subset of 1D torsions will be expanded in the future to include:
- Torsions within flexible and non-flexible rings (subject to energy upper limit);
- Non-equivalent torsions on a center bond (e.g. C-C-C-C and C-C-C-H in aromatic ring);
- Carefully selected 2D torsions.
Currently, only 1D torsions are being computed because the price of 2D scans is 10-20x higher than for 1D scans. However, we plan to expand datasets both to include 2D torsions from the “Roche set” of molecules and more fragments from other molecules not currently included in the “Roche set”. The torsion drive data appears to ForceBalance as lists of data points consisting of coordinates, energies, and gradients; thus torsion drives of any dimension can be used when fitting force field parameters.
The pipeline used to compute these torsions is QCArchive → TorsionDrive → geomeTRIC → Psi4. The selected level of theory used to generate QM torsion profiles in this step was B3LYP-D3(BJ)/DZVP. The SDF files and 2D representations (pdf) of molecules in “Roche set” are available on GitHub, including all the torsion profiles calculated up to date.
Benchmarking
A level of theory used for calculation of torsional profiles needs to provide accurate optimized geometries, and preferably, accurate conformational energies (within 0.5-1 kcal/mol). It is convenient to use the same method for geometry optimization and for energy estimation, but this can be further refined to get more accurate energies. Based on the available literature [1, 2], we opted for B3LYP-D3(BJ)/DZVP for our early set of torsion calculations as a reasonable trade-off between accuracy and computational speed. However, we will perform an additional benchmarking study for torsion drive using 15 carefully selected torsions from the set of 820 mentioned above. We have also performed a quick preliminary study looking into basis sets of similar size to DZVP, particularly because some of the bases do not have auxiliary functions for density fitting. We found that the lack of an optimized JKFit basis for DZVP and 6-31G* does not significantly impact performance, while def2-TZVP is more expensive by a factor of ~4.
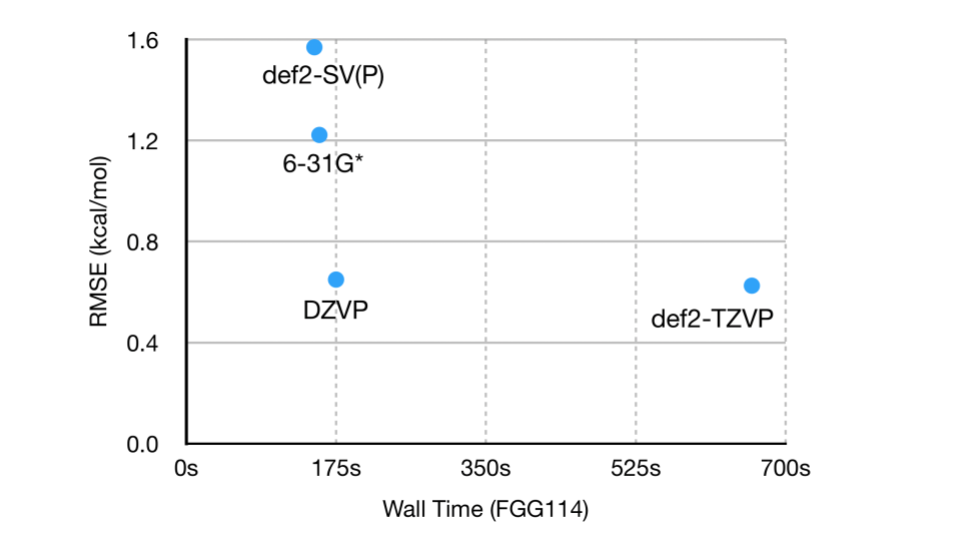
We are planning to perform benchmarking study using the following models on the selected 15 torsions. Models may include combinations of DFT functionals and basis sets, as well as semiempirical models and machine learning potentials. Based on the results of this benchmarking study, a method for future computations for parameterization sprints will be selected.