Torsion subgroup update
Posted on 16 Oct 2018 by Lee-Ping Wang
Participants
John Chodera (MSKCC), Jessica Maat (Mobley group), David Mobley (UC Irvine), Levi Naden (MolSSI), Jessica Nash (MolSSI), Yudong Qiu (Wang group), Daniel Smith (MolSSI), Chaya Stern (Chodera group), Jeff Wagner (OpenFF), Lee-Ping Wang (UC Davis)
Glossary
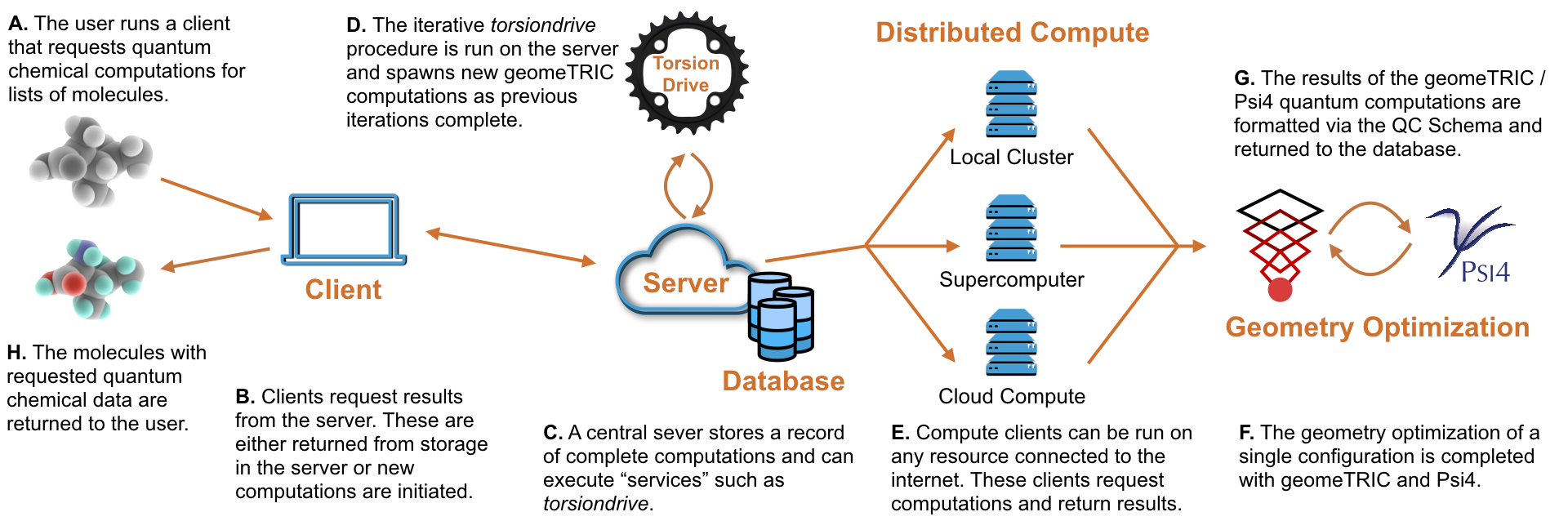
geomeTRIC: Geometry optimization software package, Wang lab
fragmenter: Fragment molecules for QM torsion scans, Chodera lab. Provides input to torsiondrive and QCFractal.
torsiondrive: Manages geometry optimization calculations for torsion scan, Wang lab. Uses geomeTRIC to carry out optimizations.
QCFractal: Distributed compute and database platform for quantum chemistry, MolSSI (mainly Daniel Smith). Part of the QCArchive ecosystem (previously called QCDB or Quantum Chemistry Database). Uses torsiondrive code to provide this calculation as a service.
cmiles: canonical SMILES string generator, Chodera lab.
Lee-Ping shared that geomeTRIC has been updated to remove all instances of numpy.matrix. Going forward, geomeTRIC should hopefully not generate any more deprecation warnings when called by torsiondrive or QCFractal.
Chaya shared updates. fragmenter is now integrated with QCFractal - it defines a workflow and registers it to the database, then generates the JSON specs for QCFractal to carry out the torsion scans. cmiles now generates 5 different flavors of SMILES strings - and hope that it will support universal SMILES soon. Also shared a document that describes the full torsion drive workflow. The basic steps are:
- Start with a list of SMILES strings representing molecules taken from a database such as DrugBank, potentially thousands or more.
- Enumerate all possible ionization, tautomers and protonation states with fragmenter.
- Determine the possible molecular fragments with fragmenter.
- Determine the torsions that need to be scanned and the corresponding atom indices.
- Scan over the torsions using torsiondrive and geomeTRIC.
- Obtain optimized xyz coordinates on the torsion grid points, corresponding energies / gradients and bond orders.
Lee-Ping mentioned there was a need for a different workflow to carry out only step (5), aka the “simplified torsion drive”. John said the “simplified torsion drive” should be a different workflow than the “full torsion drive”.
At this point, Daniel said that the “simplified torsion drive” was a service provided by QCFractal and the “full torsion drive” (including steps 1-4, i.e. fragmentation etc.) were not part of the service. While steps 1-4 could conceivably be included to make a new service, the main idea is that services are hard-coded workflows in QCFractal, and custom workflows are not easy to support.
After some discussion, it became clear that QCFractal will handle calculation and deposition of results for required QM calculations, but the preparatory steps are not within its scope so we’ll need to pay separate attention to automating a workflow to handle preparation of molecules in steps 1-4 and spawning off calculations to QCFractal in steps 5-6. Chaya’s fragmenter code already handles steps 1-4, but in a serial manner. Turning this into a scalable workflow will likely fall to Jeff Wagner.
In the remainder of the meeting, Chaya said that the optimal fragmentation scheme was still being worked on, John asked Daniel to add functions to QCFractal to provide Wiberg bond orders for optimized geometries, and Lee-Ping agreed to produce a schema for computing electrostatic potentials on grids using QCFractal.
