Benchmark: OpenFF performance on small molecule energies and geometries
Posted on 25 Jul 2020 by David Hahn
The Open Force Field Initiative, focusing on open tools and science to improve accuracy of molecular modeling, has now been up and running for approximately 18 months. Recent highlights have included the release of a series of updates to Parsley, a small molecule force field for biomolecular simulation, which is now at version OpenFF-1.2.0. Parsley was iteratively optimized to improve agreement with quantum chemical geometries, energetics, and vibrational frequencies.
Here, we provide a preliminary benchmark assessment of several small molecule force fields (FF) compared to quantum mechanical (QM) results regarding gas phase energies and geometries of a set of molecule conformers.
We have compared the vacuum-minimized energies and geometries of nine small molecule force fields to quantum chemical results (B3LYP-D3BJ/DZVP) available in QCArchive. Our dataset is the OpenFF Full Optimization Benchmark 1 set, except of molecules which were used for training OpenFF-1.2.0. The set includes about 23000 conformers of a broad range of molecules. Additional methodology details are available in our recent preprint, which will be updated soon to include this new data.
Assessed force fields:
- GAFF1
- GAFF2
- MMFF94
- MMFF94S
- OPLS3e
- Smirnoff99Frosst
- Parsley, 3 versions:
### OpenFF is competitive with other small molecule force fields #### Energetic comparisons
The energies were compared by calculating the relative energy differences ($$ddE$$) between the force field (FF) and quantum mechanical (QM) energies of a conformer $$i$$, relative to the lowest QM conformer 0:
$$ddE_i = dE_{FF,i}-dE_{QM,i}=(E_{FF,i}-E_{FF,0})-(E_{QM,i}-E_{QM,0})$$
For an ideal force field, the $$ddE$$ of all conformers would be zero, meaning the FF reproduces correctly the QM relative energies between conformers. The following graph shows the $$ddE$$ distributions for the set of conformers, as obtained for the four latest force fields of each family (other FF curves left out for clarity):
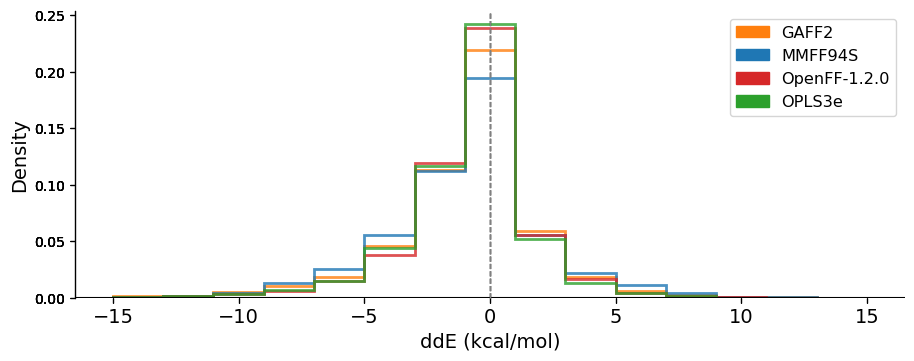
Geometric comparisons
We compared the conformer geometries between FF and QM conformers by the atom-positional root mean squared deviation (RMSD) and the torsional fingerprint deviation (TFD). The RMSD and TFD distributions are shown below:
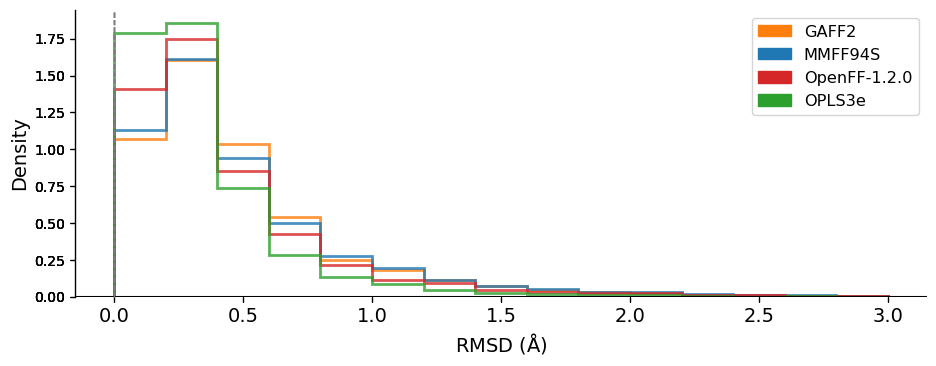
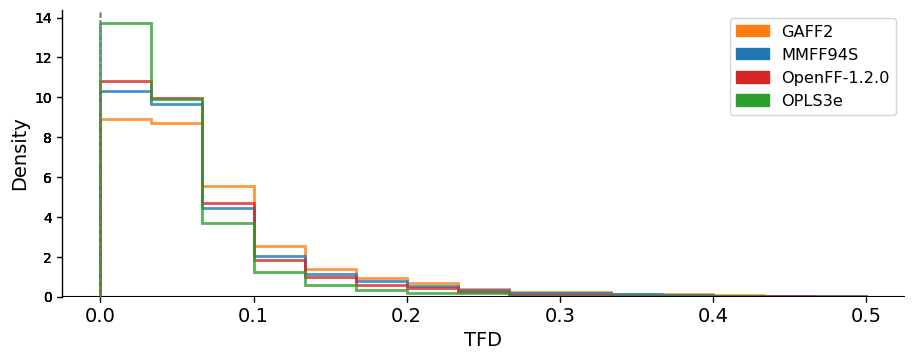
This data suggests that, at least on this dataset and judged by these metrics, OpenFF 1.2 is modestly superior to MMFF94S and GAFF2 and lags only slightly behind OPLS3e.
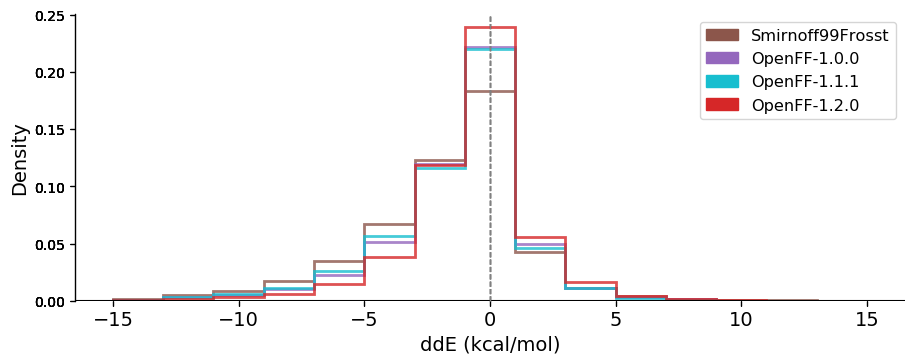
### OpenFF has progressively improved across versions The following graphs are showing the $$ddE$$ and RMSD distributions of four OpenFF versions (Smirnoff99Frosst, OpenFF-1.0.0, OpenFF-1.1.1 and OpenFF-1.2.0). They are illustrating how the performance of OpenFF improves with every version.
Energetic comparisons
Geometric comparisons
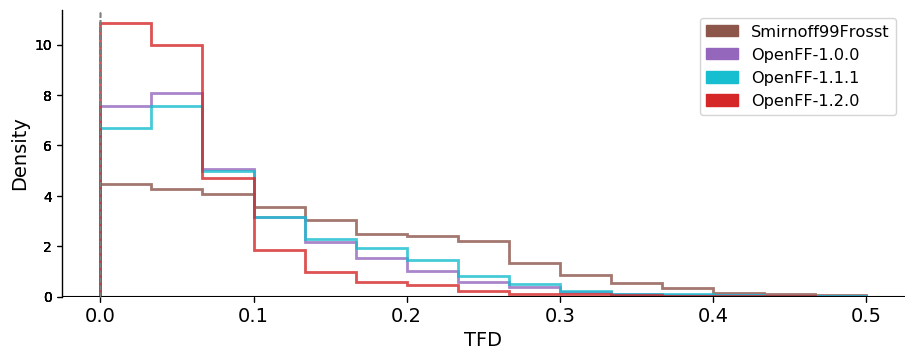
Both energies and geometries clearly improved in reproducing the QM reference across these versions, with the largest changes being from smirnoff99Frosst (our starting point force field) to OpenFF 1.0, and from OpenFF 1.0/1.1 to OpenFF 1.2 – both providing marked increases in accuracy. The tails of the distributions of the latest OpenFF version are strongly reduced compared to the OpenFF predecessor Smirnoff99Frosst (OpenFF-1.2.0 has basically no conformer deviating more than 2 Angstroms from QM on this dataset).
Results suggest that OpenFF-1.2.0 is the best “public” force field available now. Among the force fields tested and based on the investigated properties, OpenFF-1.2.0 ranks second after OPLS3e. It is therefore one of the best public general small molecule force fields for biomolecular simulations available, at least based on performance on this dataset Future releases of the Open Force Field will improve further The iterative improvement of Parsley is ongoing work. Future releases will have increased accuracy and a broader molecule coverage by adding the following features:
- Wiberg Bond Order (WBO) torsion interpolation
- Lennard Jones parameter refitting
- Off-site charges
- Bespoke parameters
- Refinement and extension of reference data sets
Furthermore additional benchmarking studies will be published targeting liquid phase properties, general host-guest and more specifically protein-ligand interactions.
Further reading
Further information about Open Force Field’s parameterization and benchmarking strategies is presented in this blog post. More details about the present study can be found in this preprint.
Contributions
These people have contributed to this benchmark assessment:
- Victoria Lim (UC Irvine)
- David Hahn (Janssen R&D)
- Gary Tresadern (Janssen R&D)
- Christopher Bayly (OpenEye Scientific)
- David Mobley (UC Irvine)
